Structures of macromolecular machines
Research interests
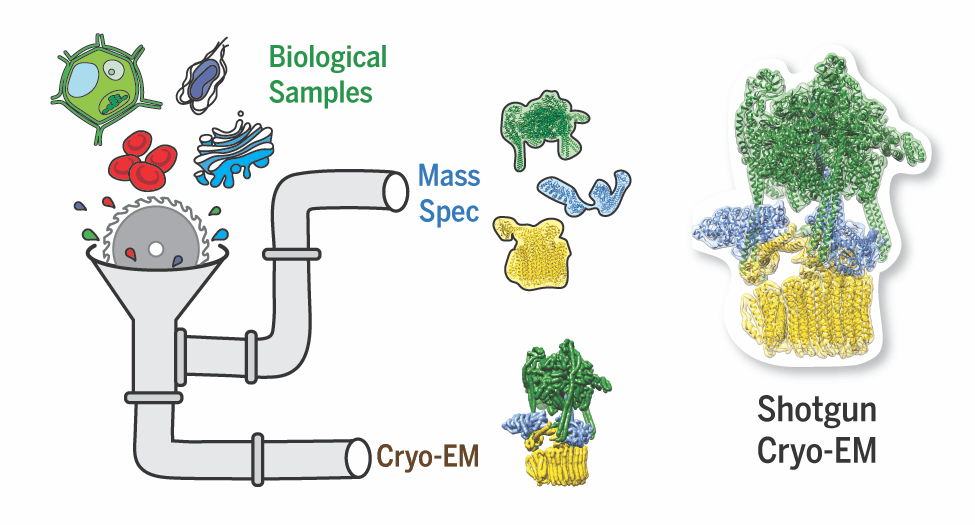

Multi-protein complexes are necessary for nearly all cellular processes, and understanding their structure is required for elucidating their function. Current high-resolution strategies in structural biology are effective but lag behind other fields (e.g., genomics and proteomics) due to their reliance on purified samples rather than heterogeneous mixtures. We are working on a method combining single-particle analysis by electron microscopy with protein identification by mass spectrometry to structurally characterize macromolecular complexes called (shotgun EM).
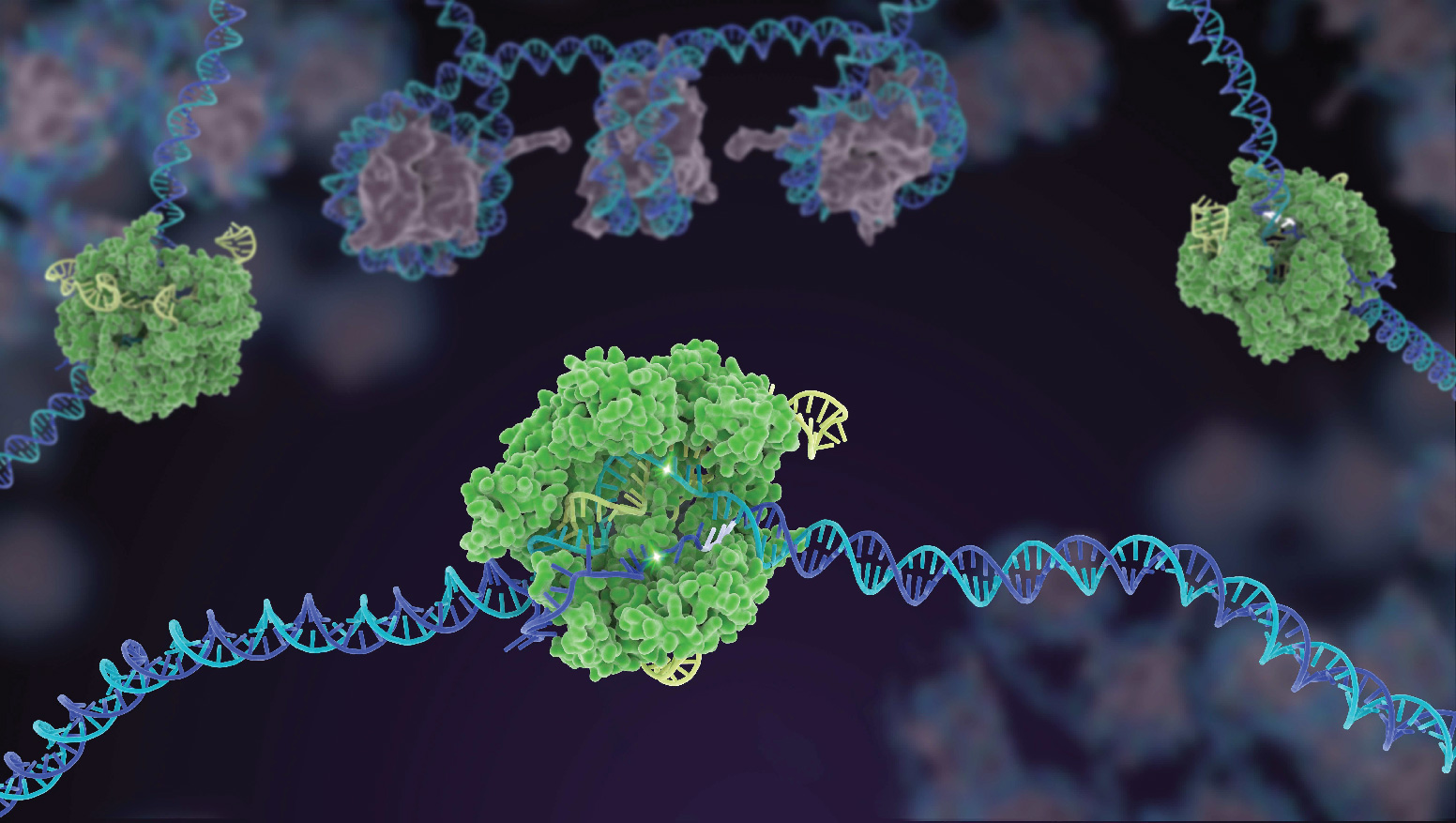
We use cryogenic electron microscopy (cryo-EM) to determine high-resolution structures of multi-subunit CRISPR effector complexes. Our structures provide critical insights into the molecular mechanisms underlying assembly, activation and regulation of these prevalent yet poorly-understood complexes. Our structural models can then serve as a blueprint for developing a the next generation of Cascade-based tools for precision genome engineering.
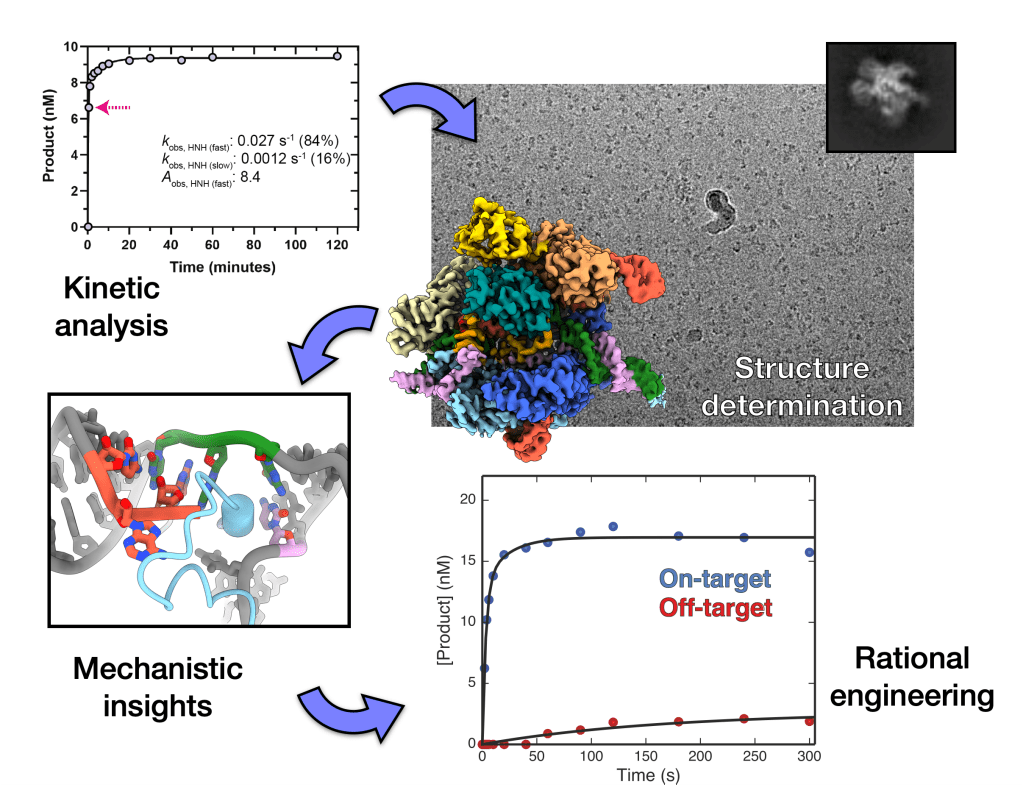
We are developing methods that use precise kinetic analysis to guide the selection of substrates and time-points for structure determination using cryo-EM. This approach allows us to directly visualise mechanistically important molecular intermediates of dynamic complexes. The transient conformational intermediates can then be used to guide rational redesign of biomolecular machines with improved enzymatic activities and altered .
Get the latest content from the Taylor Lab.
All content © David Taylor, Dustin Taylor, Candice Chen, Jack Bravo 2025
